- Background
- Neovim-based IDE for R
- Parallel R with batchtools
- References
December 18, 2020
Outline
R Language
About
- Statistical environment and programming language (CRAN) widely used in academia and data science.
- Free and runs on all common operating systems
- Large ecosystem of extension packages, e.g. Bioconductor and CRAN
Working environments
- R console and basic Rguis
- RStudio
- Jupyter Notebooks and JupyterHub
- Nvim-R-tmux: Neovim-based IDE for R
- Emacs Speaks Statistics (ESS)
- Many additional options: Rgedit, RKWard, Eclipse, Tinn-R, Notepad++, NppToR
RStudio Server for Web-based HPCC Access
- Integrated development environment (IDE) for R. RStudio local GUI and RStudio Server is web-based.
- User access to RStudio Server on HPCC
- The URL is: https://rstudio.hpcc.ucr.edu
- Username and password: same as for ssh access to HPCC
Some useful RStudio shortcuts:
Ctrl+Enter: send code to R consoleCtrl+Alt+Enter: send code to terminalCtrl+Shift+C: comment/uncommentCtrl+1/2: switch window focus
Nvim-R-Tmux: Terminal-based R Environment
URLs: 1. main page, 2. HPCC manual, 3. custom installs
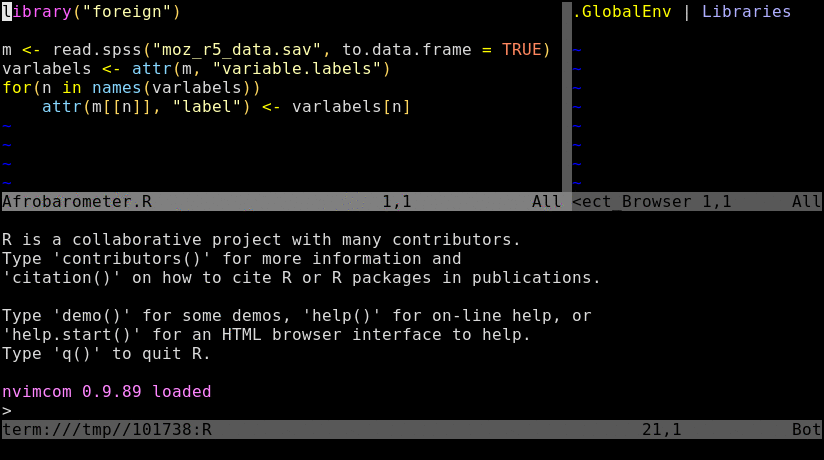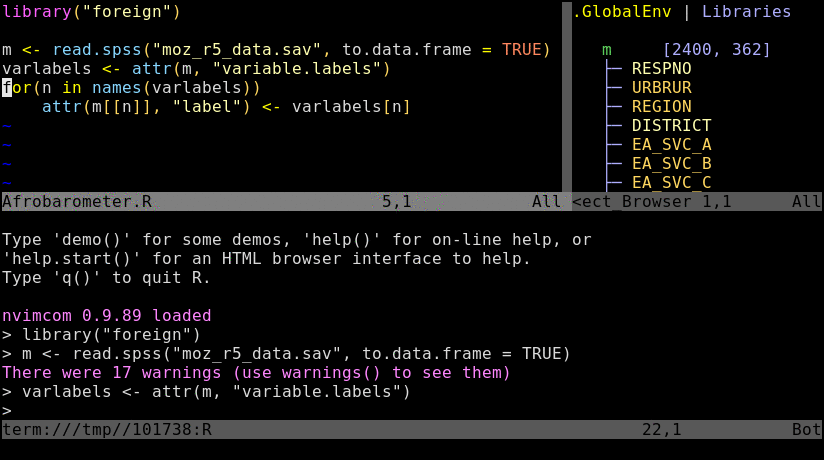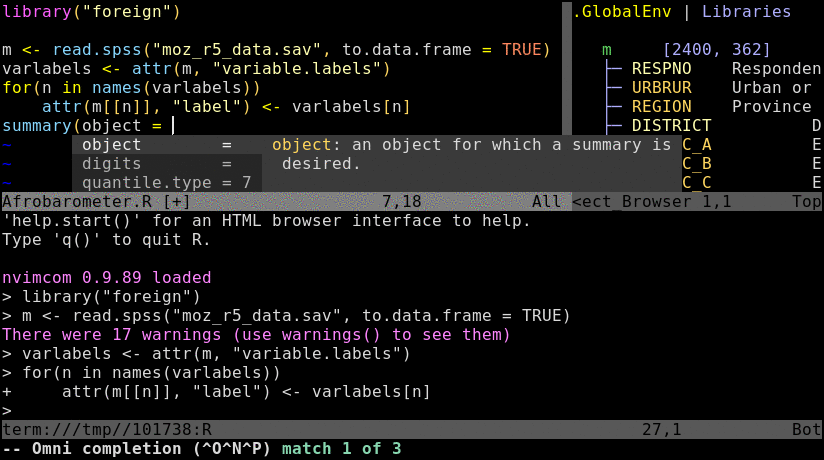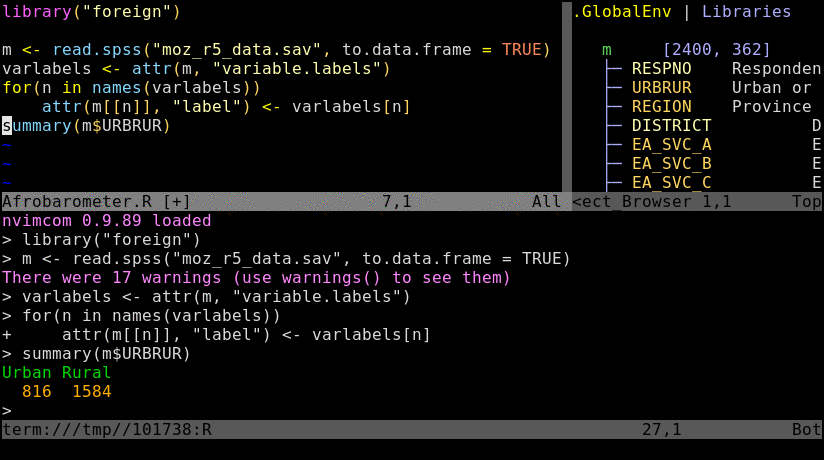
Animated screenshot of Nvim-R (from here)
Advantages of Command-line UI
- Knowledge of command-line interface is essential for working on a computer cluster efficiently
- Main advantage: language agnostic approach that works with most computer languages
- Users of Emacs may want to consider using ESS instead
Outline
- Background
- Neovim-based IDE for R
- Parallel R with batchtools
- References
Introduction to Nvim-R-Tmux
- The following introduces Nvim-R combined with Tmux.
- Similar instructions are available in HPCC’s Nvim-R-Tmux tutorial here.
- Note: Nvim-R and Tmux are two independent tools that can be used independently or in combination, and both are useful for remote terminal work.
- For simplicity, some of the following examples use Nvim-R without Tmux. Once users know the basics of both then it is trivial to combine them as needed.
Vim/Nvim Basics
The following opens a file (here myfile.txt) with nvim (or vim). This can be a new file or an existing one.
nvim myfile.txt # for neovim (or 'vim myfile.txt' for vim)
Modes
In Vim/Nvim there are three main modes: normal, insert and command mode. The most important commands for switching between the three modes are:
i: switches from the normal to the insert mode. The latter is used for typing.Esc: switches from the insert mode back to the normal mode.:: starts the command mode (from normal mode) at the bottom of the terminal window.
The cursor is moved with the arrow keys. In Nvim one can also enable mouse-based movements of the cursor. Fn Up/Down allows to page. In the following, all commands starting with : need to be typed in the command mode. All other commands are typed in the normal mode after pushing the Esc key.
[ Scroll down to continue ]
Important modifier keys to control Vim/Nvim
:w: saves changes to file. If in editing mode,Escneeds to be pressed first.:q: quits file that has not been changed; useq!to quit file without saving changes.:wq: saves and quits file
Useful resources for learning Vim/Nvim
Tmux for Managing Terminal Sessions
What is Tmux?
- Tmux is a virtual terminal multiplexer providing re-attachable terminal sessions
- Advantage: work in a terminal session cannot get lost due to internet disruptions or even when switching computers
- Combined with the
Nvim-rplugin it provides a flexible working environment for R - Users can send code from a script to the R console or command-line.
- On HPCC both Nvim-R and Tmux are pre-configured and easy to install

Nvim-R-Tmux Configuration in HPCC User Accounts
Skip these steps if Nvim-R-Tmux is already configured in your account. Or follow the detailed instructions to install Nvim-R-Tmux from scratch on your own system (e.g. laptop or computer).
1. Log in to your user account on HPCC and execute on the command-line:
install_nvimRtmux
2. To enable the nvim-R-tmux environment, log out and in again.
3. Follow usage instructions of next section.
Typical Usage Workflow for Nvim-R-Tmux
1. Start tmux session from login node (not compute node!)
Running Nvim from tmux provides reattachment functionality. Skip this step if this is not required.
tmux # starts a new tmux session tmux a # attaches to an existing session
2. Open nvim-connected R session
Open a *.R or *.Rmd file with nvim and initialize a connected R session with \rf. Note, the resulting split window among Nvim and R behaves like a split viewport in nvim or vim meaning the usage of Ctrl-w w followed by i and Esc is important for session navigation.
nvim myscript.R # or *.Rmd file
3. Send R code from nvim to the R pane
Single lines of code can be sent from nvim to the R console by pressing the space bar. To send several lines at once, one can select them in nvim’s visual mode and then hit the space bar.
[ Scroll down to continue ]
- Please note, the default command for sending code lines in the nvim-r-plugin is
\l. This key binding has been remapped in the provided.config/nvim/init.vimfile to the space bar. Most other key bindings (shortcuts) still start with the\as LocalLeader, e.g.\rhopens the help for a function/object where the cursor is located in nvim. More details on this are given on the next slide(s). - The most comprehensive manual on this is the official
Nvim-Rdocumentation here.
Keybindings to Control Environment
Important keybindings for nvim
\rf: opens vim-connected R session. If you do this the first time in your user account, you might be asked to create anRdirectory under~/. If so approve this action by pressingy.spacebar: sends code from vim to R; here remapped ininit.vimfrom default\l:splitor:vsplit: splits viewport (similar to pane split in tmux)gz: maximizes size of viewport in normal mode (similar to Tmux’sCtrl-a zzoom utility)Ctrl-w w: jumps cursor to R viewport and back; toggle between insert (i) and command (Esc) mode is required for navigation and controlling the environment.Ctrl-w r: swaps viewportsCtrl-w =: resizes splits to equal size:resize <+5 or -5>: resizes height by specified value
[ Scroll down to continue ]
:vertical resize <+5 or -5>: resizes width by specified valueCtrl-w HorCtrl-w K: toggles between horizontal/vertical splitsCtrl-spacebar: omni completion for R objects/functions when nvim is in insert mode. Note, this has been remapped ininit.vimfrom difficult to type defaultCtrl-x Ctrl-o.:h nvim-R: opens nvim-R’s user manual; navigation works the same as for any Vim/Nvim help document:Rhelp fct_name: opens help for a function from nvim’s command mode with text completion supportCtrl-s and Ctrl-x: freezes/unfreezes vim (some systems)
Important keybindings for tmux
Pane-level commands
Ctrl-a %: splits pane verticallyCtrl-a ": splits pane horizontallyCtrl-a o: jumps cursor to next paneCtrl-a Ctrl-o: swaps panesCtrl-a <space bar>: rotates pane arrangementCtrl-a Alt <left or right>: resizes to left or rightCtrl-a Esc <up or down>: resizes to left or right
Window-level comands
Ctrl-a n: switches to next tmux windowCtrl-a Ctrl-a: switches to previous tmux windowCtrl-a c: creates a new tmux windowCtrl-a 1: switches to specific tmux window selected by number
Session-level comands
Ctrl-a d: detaches from current sessionCtrl-a s: switch between available tmux sessions$ tmux new -s <name>: starts new session with a specific name$ tmux ls: lists available tmux session(s)$ tmux attach -t <id>: attaches to specific tmux session$ tmux attach: reattaches to session$ tmux kill-session -t <id>: kills a specific tmux sessionCtrl-a : kill-session: kills a session from tmux command mode
Use Same Environment for Other Languages
Basics
For languages other than R one can use the vimcmdline plugin for nvim (or vim). Supported languages include Bash, Python, Golang, Haskell, JavaScript, Julia, Jupyter, Lisp, Macaulay2, Matlab, Prolog, Ruby, and Sage. The nvim terminal also colorizes the output, as in the screenshot below, where different colors are used for general output, positive and negative numbers, and the prompt line.

Install
To install it, one needs to copy from the vimcmdline repository the directories ftplugin, plugin and syntax and their files to ~/.config/nvim/. For user accounts of UCR’s HPCC, the above install script install_nvimRtmux includes the install of vimcmdline (since 09-Jun-18).
Usage
The usage of vimcmdline is very similar to nvim-R. To start a connected terminal session, one opens with nvim a code file with the extension of a given language (e.g. *.sh for Bash or *.py for Python), while the corresponding interactive interpreter session is initiated by pressing the key sequence \s (corresponds to \rf under nvim-R). Subsequently, code lines can be sent with the space bar. More details are available here.
Nvim-R Demo
To try out the following instructions, users want to log into their HPCC account via ssh, and then preferentially connect to a node by initializing an interactive srun session. The latter mimics the best practices for a real workflow but is not necessary for this basic exercise.
srun --x11 --partition=short --mem=2gb --cpus-per-task 4 --ntasks 1 --time 1:00:00 --pty bash -l
- Under
--partitionit is important to assign the name of a partition a user has access to- Most users have access to:
short,batch,intelandhighmem - Students registered under the statistics department, only have access to:
statsdept - Users of labs owning computer nodes also can access:
<pi_name>lab
- Most users have access to:
- For more details on argument settings for
srun, see here
Download R_for_HPC_demo.R file to you HPCC account as follows.
wget https://raw.githubusercontent.com/ucr-hpcc/ucr-hpcc.github.io/master/presentations/2020-12-18_Workshop/R_for_HPC/demo_files/R_for_HPC_demo.R
[ Scroll down to continue ]
Open nvim_demo.R with nvim. The content of this file is shown in the following code block. Next, initialize a Nvim-connected R session with \rf, and then execute the code by pressing the space bar on your keyboard.
library(tidyverse)
write_tsv(iris, "iris.txt") # Creates sample file
read_tsv("iris.txt") %>% # Import with read_tbv from readr package
as_tibble() %>%
group_by(Species) %>%
summarize_all(mean) %>%
reshape2::melt(id.vars=c("Species"), variable.name = "Samples", value.name="Values") %>%
ggplot(aes(Samples, Values, fill = Species)) +
geom_bar(position="dodge", stat="identity")
[ Scroll down to continue ]
If X11 is enabled in a user session then the above code will generate the following bar plot in a separate graphics window.
Selecting R Versions on HPCC
- Like many other software tools, R versions are managed under HPCC’s module system.
- To use a specific R version in Nvim-R, one simply loads it prior to starting Nvim. Instructions for enabling additional R version toggle options are provided here.
Which R versions are available can be listed with the following command.
module avail R
The version labeled default is used by default. A specific R version can be loaded as follows.
module load R/4.0.1
Check which modules (including R) are loaded in a user’s environment.
module list
Outline
- Background
- Neovim-based IDE for R
- Parallel R with batchtools
- References
Parallel Evaluations in R
- R provides a large number of packages for parallel evaluations on multi-core, multi-socket and multi-node systems. The latter are usually referred to as computer clusters.
- MPI is also supported
- For an overview of parallelization packages available for R see here
- One of the most comprehensive parallel computing environments for R is
batchtools. Older versions of this package were released under the nameBatchJobs(Bischl et al. 2015). batchtoolssupports both multi-core and multi-node computations with and without schedulers. By making use of cluster template files, most schedulers and queueing systems are supported (e.g. Torque, Sun Grid Engine, Slurm).
Reminder: Traditional Job Submission for R
This topic is covered in more detail in other tutorials. The following only provides a very brief overview of this submission method.
1. Create Slurm submission script, here called script_name.sh with:
#!/bin/bash -l #SBATCH --nodes=1 #SBATCH --ntasks=1 #SBATCH --cpus-per-task=1 #SBATCH --mem-per-cpu=1G #SBATCH --time=1-00:15:00 # 1 day and 15 minutes #SBATCH --mail-user=useremail@address.com #SBATCH --mail-type=ALL #SBATCH --job-name="some_test" #SBATCH -p short # Choose queue/partition from: intel, batch, highmem, gpu, short Rscript my_script.R
2. Submit R script (my_script.R) called by above Slurm script with:
sbatch script_name.sh
Parallel Evaluations on Clusters with batchtools
- The following introduces the usage of
batchtoolsfor a computer cluster using SLURM as scheduler (workload manager). SLURM is the scheduler used by the HPCC. - Similar instructions are provided in HPCC’s manual section covering
batchtoolshere - To simplify the evaluation of the R code on the following slides, the corresponding text version is available for download from here.
Hands-on Demo of batchtools
Set up working directory for SLURM
First login to your cluster account, open R and execute the following lines. This will create a test directory (here mytestdir), redirect R into this directory and then download the required files:
dir.create("mytestdir")
setwd("mytestdir")
download.file("https://bit.ly/3gZJBsy", "slurm.tmpl")
download.file("https://bit.ly/3nvSNHA", ".batchtools.conf.R")
Load package and define some custom function
The following code defines a test function (here myFct) that will be run on the cluster for demonstration purposes.
[ Scroll down to continue ]
The test function (myFct) subsets the iris data frame by rows, and appends the host name and R version of each node where the function was executed. The R version to be used on each node can be specified in the slurm.tmpl file (under module load).
library('RenvModule')
module('load','slurm') # Loads slurm among other modules
library(batchtools)
myFct <- function(x) {
Sys.sleep(10) # to see job in queue, pause for 10 sec
result <- cbind(iris[x, 1:4,],
Node=system("hostname", intern=TRUE),
Rversion=paste(R.Version()[6:7], collapse="."))
}
Submit jobs from R to cluster
The following creates a batchtools registry, defines the number of jobs and resource requests, and then submits the jobs to the cluster via SLURM.
reg <- makeRegistry(file.dir="myregdir", conf.file=".batchtools.conf.R") Njobs <- 1:4 # Define number of jobs (here 4) ids <- batchMap(fun=myFct, x=Njobs) done <- submitJobs(ids, reg=reg, resources=list(partition="short", walltime=120, ntasks=1, ncpus=1, memory=1024)) waitForJobs() # Wait until jobs are completed
Summarize job status
After the jobs are completed one can inspect their status as follows.
getStatus() # Summarize job status showLog(Njobs[1]) # killJobs(Njobs) # # Possible from within R or outside with scancel
Access/assemble results
The results are stored as .rds files in the registry directory (here myregdir). One can access them manually via readRDS or use various convenience utilities provided by the batchtools package.
readRDS("myregdir/results/1.rds") # reads from rds file first result chunk
loadResult(1)
lapply(Njobs, loadResult)
reduceResults(rbind) # Assemble result chunks in single data.frame
do.call("rbind", lapply(Njobs, loadResult))
Remove registry directory from file system
By default existing registries will not be overwritten. If required one can explicitly clean and delete them with the following functions.
clearRegistry() # Clear registry in R session
removeRegistry(wait=0, reg=reg) # Delete registry directory
# unlink("myregdir", recursive=TRUE) # Same as previous line
Load registry into R
Loading a registry can be useful when accessing the results at a later state or after moving them to a local system.
from_file <- loadRegistry("myregdir", conf.file=".batchtools.conf.R")
reduceResults(rbind)
Conclusions
Nvim-R-Tmux
- Steeper learning curve than GUI-based IDEs, including RStudio or Jupyter Notebooks
- However, it is much more
- powerful, flexible, robust and language agnostic solution for working on remote systems
- time learning it is well invested, especially for students and researchers with complex data analysis and programming needs
Advantages of batchtools
- many parallelization methods multiple cores, and across both multiple CPU sockets and nodes
- most schedulers supported
- takes full advantage of a cluster
- robust job management by organizing results in registry file-based database
- simplifies submission, monitoring and restart of jobs
- well supported and maintained package
Outline
- Background
- Neovim-based IDE for R
- Parallel R with _batchtools
- References
References
Bischl, Bernd, Michel Lang, Olaf Mersmann, Jörg Rahnenführer, and Claus Weihs. 2015. “BatchJobs and BatchExperiments: Abstraction Mechanisms for Using R in Batch Environments.” Journal of Statistical Software. http://www.jstatsoft.org/v64/i11/.